1. Molecular Plant Pathology
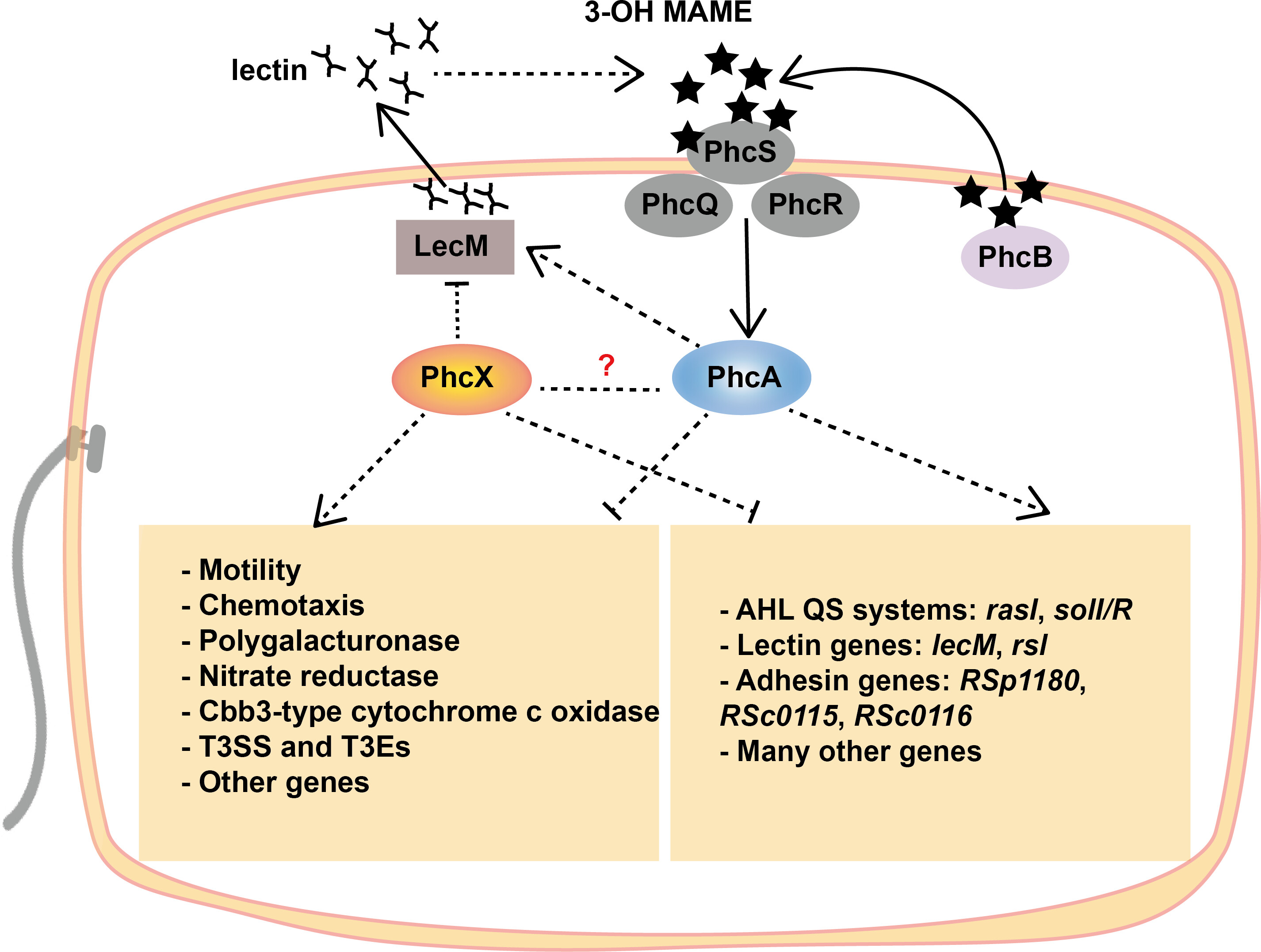
Our lab is fundamentally driven by understanding the molecular dialogue between plants and microbes. A primary focus is Molecular Plant Pathology, where we decipher the genetic mechanisms underlying host specificity, pathogenicity, and virulence. Using the Ralstonia solanacearum species complex and the litchi-specific oomycete Peronophytophthora litchii as model systems, we employ integrated multi-omics and functional genetics to unravel regulatory networks (e.g., quorum-sensing), characterize key virulence factors (like Type III and VI effectors), and elucidate the critical molecular events that define host-pathogen interactions. Our goal is to translate this knowledge into novel strategies for managing devastating plant diseases. (Liu et al. 2023, mBio).
2. Development and Utilization of Microbial Resources
Complementing this focus on pathogenesis is our work on the Development and Utilization of Microbial Resources. We actively explore microbial biodiversity to discover and harness beneficial microorganisms. This involves the systematic isolation and screening of bacteria and fungi from various environments to identify potent biocontrol agents against phytopathogens. Furthermore, our discovery pipeline often leads to the identification and taxonomic characterization of novel bacterial species, adding to the fundamental understanding of microbial diversity and opening new avenues for agricultural and biotechnological applications.
3. Development of Bioinformatic Tools and Algorithms
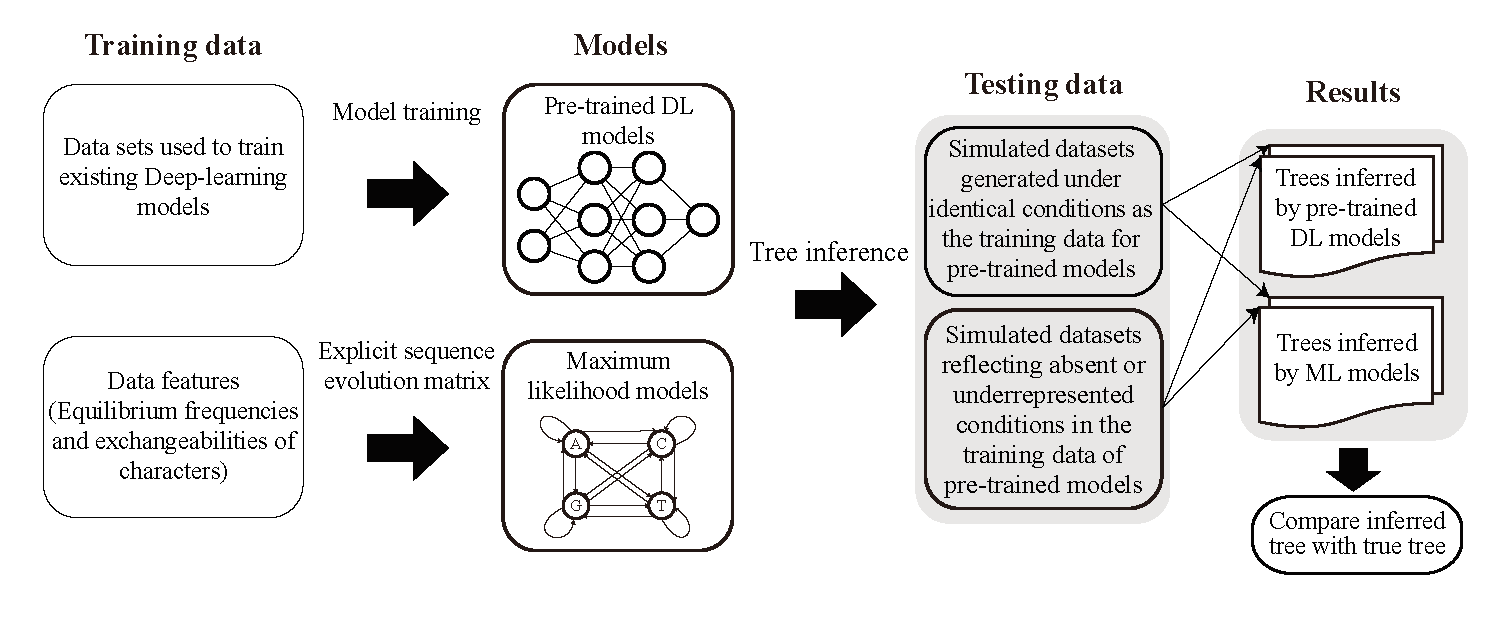
Underpinning all our wet-lab research is the Development of Bioinformatic Tools and Algorithms. We are committed to building computational capacity to accelerate biological discovery. Our lab creates and applies innovative software and algorithms to tackle specific challenges in genomics, such as processing complex multi-omics data from our pathogen studies, facilitating the discovery of secondary metabolites in biocontrol strains, and reconstructing robust phylogenetic trees to understand evolutionary relationships. These tools are designed to not only serve our internal projects but also to be shared with the broader scientific community to advance research in molecular phylogenetics and beyond.